Introduction
Contamination of TS-like cells detected in RNA-seq data from FI-SC
Dr. Endo analysed the RNA-seq dataset associated with the original STAP papers and found that FI stem cell (FI-SC) samples were the mixture of 2 different cell types which have distinct expression profiles [1]. His findings include:
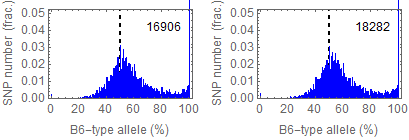
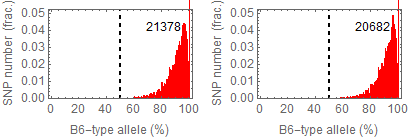
- Only FI-SC samples have a skewed allele frequency distribution whose peak is at around 95% B6-type allele
- Only FI-SC samples have different allele heterozygosity depending on gene types; their TSC-specific genes have less B6 homo SNPs and conversely ESC-specific genes have more B6 homo SNPs
- Most of the TSC-specific genes in FI-SC samples are expressed in approxmately 10% of TSC samples in the same dataset
Here I reproduced most of the figures in Figure 2 and Figure 4 in Dr. Endo's paper and confirmed his results.
Note: I am a beginner for NGS data analysis (about 3 month experience). The result presented in this page is only preliminary and in non-prefessional quality.
1st. trial
For data retrieval and software setup/usage, see Trisomy-8 detection.Mapping
It took 7-9h when using 1 CPU core for TruSeq sequences below. I could run 2 mappings (rep1 and rep2) at the same time.
| Accesion | Name | accepted_hits.bam | Index of .bam | align_summary.txt |
|---|---|---|---|---|
| SRR1171565 | FI-SC-rep1 | SRR1171565.bam (2.6GB) , stats. | SRR1171565.bam.bai (2MB) | align_summary.txt |
| SRR1171566 | FI-SC-rep2 | SRR1171566.bam (2.4GB) , stats. | SRR1171565.bam.bai (2MB) | align_summary.txt |
| SRR1171590 | TSC-rep1 | SRR1171590.bam (2.8GB) , stats. | SRR1171590.bam.bai (2MB) | align_summary.txt |
| SRR1171591 | TSC-rep2 | SRR1171591.bam (2.6GB) , stats. | SRR1171591.bam.bai (2MB) | align_summary.txt |
| SRR1171560 | ESC-rep1a | SRR1171560.bam (2.2GB) , stats. | SRR1171560.bam.bai (2MB) | align_summary.txt |
| SRR1171561 | ESC-rep2a | SRR1171561.bam (2.5GB) , stats. | SRR1171561.bam.bai (2MB) | align_summary.txt |
| SRR1171580 | STAP-rep1a | SRR1171580.bam (2.6GB) , stats. | SRR1171580.bam.bai (2MB) | align_summary.txt |
| SRR1171581 | STAP-rep2a | SRR1171581.bam (2.3GB) , stats. | SRR1171581.bam.bai (2MB) | align_summary.txt |
SNP analysis
$ ~/snpexp/src/snpexp -o snpexp.out -G ~/stap/genome/Mus_musculus/NCBI/GRCm38/Annotation/Genes/genes.gtf \ -V ~/stap/snps/dbSNP/mgp.v3.snps.rsIDdbSNPv137.vcf \ -m 20 -s C57BL6NJ,129P2,129S1,129S5 \ SRR1171565.bam $ python ~/afV2.py snpexp.out > snpexp.129B6.out $ grep -Fw -f Endo-fig2B-SNPs-ID.list snpexp.out > SRR1171565.fig2B.out
Allele frequency distribution
Ref: Endo-2014, Fig.2A
Allele frequencies of SNPs in total chromosome
ESC-rep1a, ESC-rep2a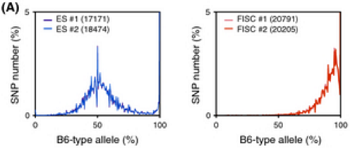
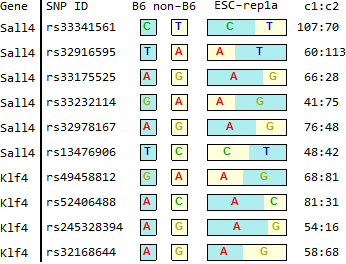
SNPs IDs used in the figures in Endo-2014:
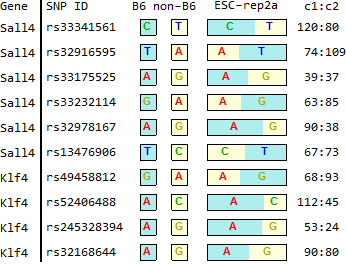
SNP allele counts (in read)
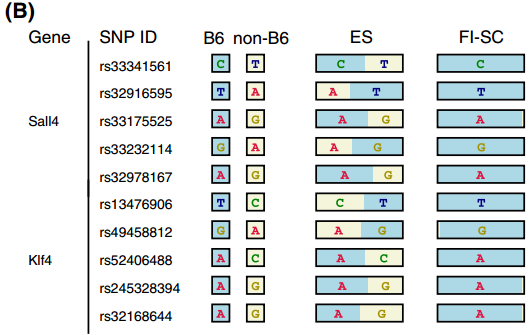
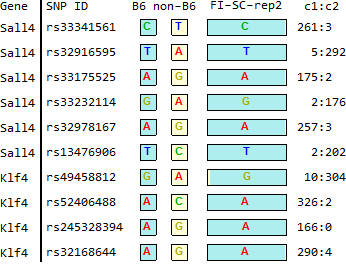
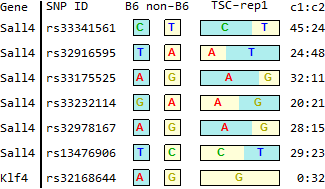
for ES-abundant genes in Endo-2014, Fig2B
Ref: Endo-2014, Fig.2B
 ESC
ESC
 FI-SC
FI-SC
 TSC
TSC
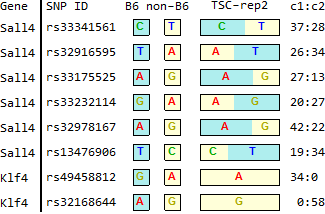
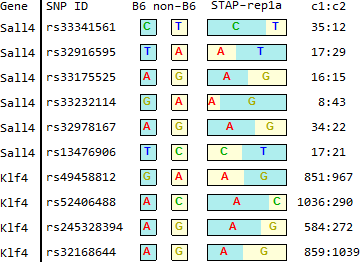 STAP
STAP
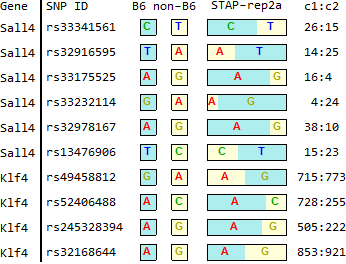
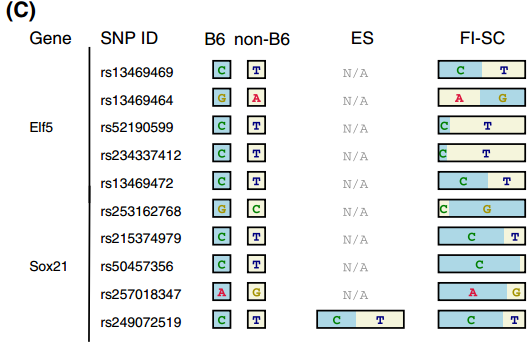
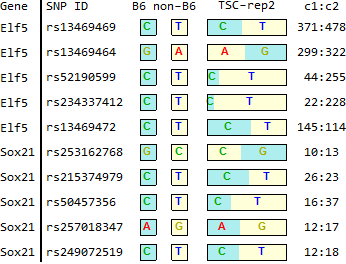
for TSC-specific genes in Endo-2014, Fig2C
Ref: Endo-2014, Fig.2C
 ESC
ESC
 FI-SC
FI-SC
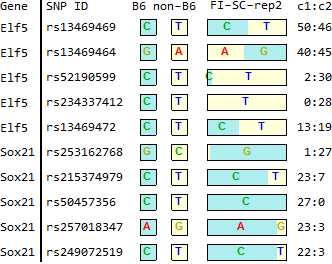
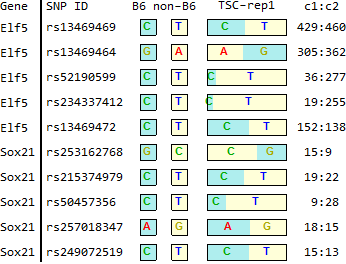 TSC
TSC
 STAP
STAP
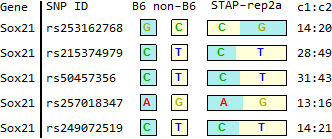
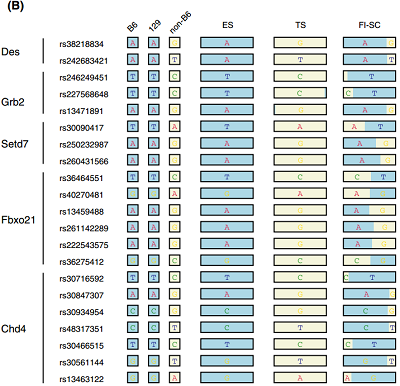
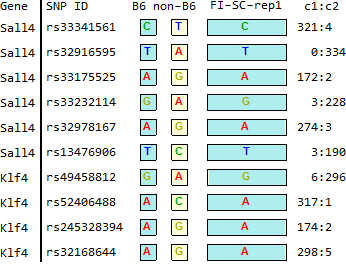
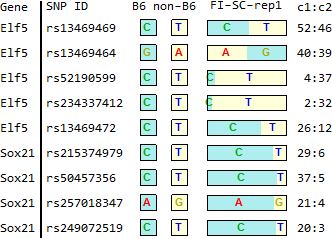
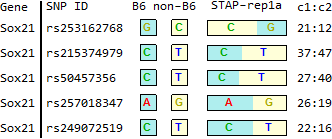
for SNPs listed in Endo-2014, Fig4B
Ref: Endo-2014, Fig.4B
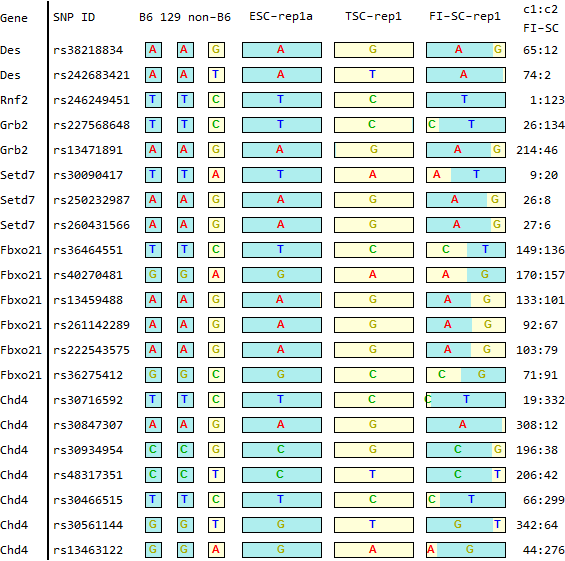
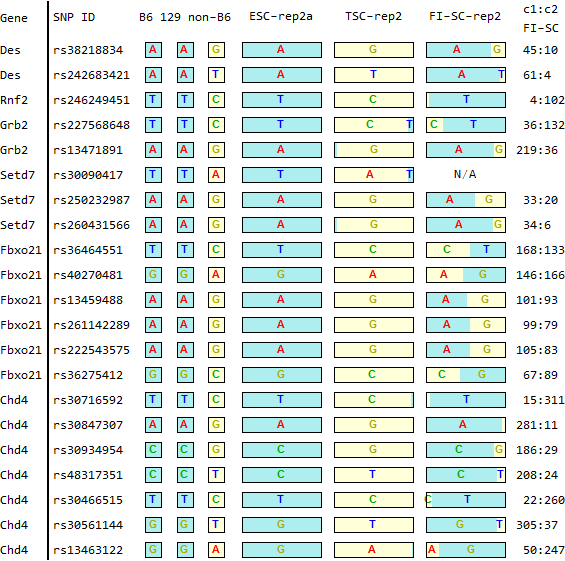
Gene expression analysis
$ ~/cufflinks-2.2.1.Linux_x86_64/cuffdiff -o truseq4.cuffdiff ../genome/Mus_musculus/NCBI/GRCm38/Annotation/Genes/genes.gtf ESC-rep1a/SRR1171560.tophat/SRR1171560.bam,ESC-rep2a/SRR1171561.tophat/SRR1171561.bam STAP-rep1a/SRR1171580.tophat/SRR1171580.bam,STAP-rep2a/SRR1171581.tophat/SRR1171581.bam FI-SC-rep1/SRR1171565.tophat/SRR1171565.bam,FI-SC-rep2/SRR1171566.tophat/SRR1171566.bam TSC-rep1/SRR1171590.tophat/SRR1171590.bam,TSC-rep2/SRR1171591.tophat/SRR1171591.bam
$ head -n1 genes.read_group_tracking > ~/fig4A.genes.read_group_tracking $ grep -Fw -f ~/Endo-Fig4A-genes.list genes.read_group_tracking >> ~/fig4A.genes.read_group_tracking
- truseq4.cuffidff/read_groups.info
- truseq4.cuffdiff/genes.read_gruop_tracking (4MB)
- truseq4.cuffdiff/genes.fpkm_tracking (8MB)
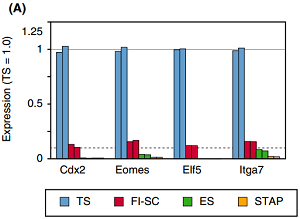
Ref: Endo-2014, Fig.4A
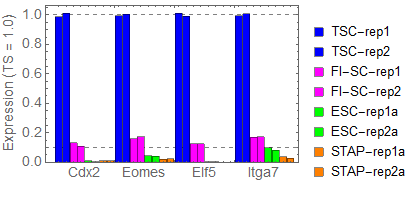
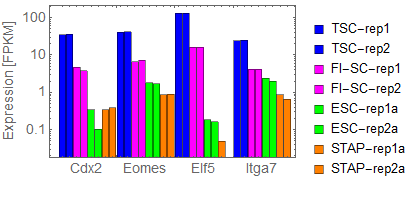
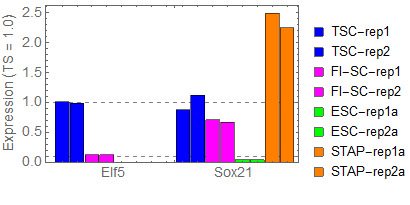
Extraction of cell-type specific genes
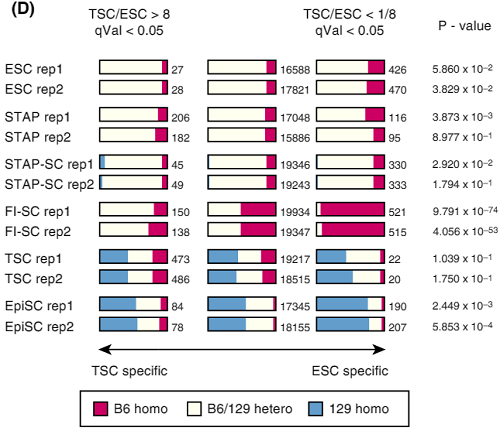
- ESC specific: q_value < 0.05 and TSC/ESC < 1/8
- TSC specific: q_value < 0.05 and TSC/ESC > 8
- others
specific-genes.py by Expo70
import sys
# extract cell-type specific genes according to Endo 2014, Fig.2D from gene_exp.diff
# by Expo70
def main():
minRPKM = 1e-2
colnum_gene = 2
colnum_sample_1 = 4
colnum_sample_2 = 5
colnum_status = 6
colnum_value_1 = 7
colnum_value_2 = 8
colnum_log2FC = 9
colnum_q_value = 12
if len(sys.argv) != 3:
print >> sys.stderr, "usage: python specific-genes.py [ES|TS|other] gene_exp.diff"
return 1
filename = sys.argv[2]
cell_type = sys.argv[1]
lineno = 0
with open(filename, "r") as fin:
for line in fin:
lineno = lineno + 1
li = line.rstrip()
ls = li.split("\t")
if lineno == 1: #check file header
labels = { colnum_gene:"gene", colnum_sample_1:"sample_1", colnum_sample_2:"sample_2", colnum_status:"status", colnum_value_1:"value_1", colnum_value_2:"value_2", colnum_log2FC:"log2(fold_change)", colnum_q_value:"q_value" }
for colnum in labels.keys():
if ls[colnum] != labels[colnum]:
print >> sys.stderr, "column '" + labels[colnum] + "' could not be found"
return 2
else:
gene = ls[colnum_gene]
sample_1 = ls[colnum_sample_1]
sample_2 = ls[colnum_sample_2]
status = ls[colnum_status]
value_1 = ls[colnum_value_1]
value_2 = ls[colnum_value_2]
if ls[colnum_log2FC] == '-inf':
log2FC = -100.0
elif ls[colnum_log2FC] == 'inf':
log2FC = 100.0
else:
log2FC = float(ls[colnum_log2FC])
q_value = float(ls[colnum_q_value])
if (status == 'OK') and (sample_1 == 'q1') and (sample_2 == 'q4'): #q1: ESC, q4: TSC
if (q_value < 0.05) and (log2FC > 3.0):
if cell_type == 'TS':
print gene
elif (q_value < 0.05) and (log2FC < -3.0):
if cell_type == 'ES':
print gene
else:
if cell_type == 'other':
print gene
if __name__ == '__main__':
main()
$ cd ~/stap/reads/truseq4.cuffdiff $ python specific-genes.py ES gene_exp.diff > es-specific-genes.list $ python specific-genes.py TS gene_exp.diff > ts-specific-genes.list $ python specific-genes.py other gene_exp.diff > other-genes.list
$ wc -l *.list
414 es-specific-genes.list 12713 other-genes.list 429 ts-specific-genes.list 13556 total
Homozygous/heterozygous compositions of SNPs for cell-type specific gene sets
$ grep -Fw -f ts-specific-genes.list snpexp.129B6.out > snpexp.cell-type-TS.out
Ref: Endo-2014, Fig.2D
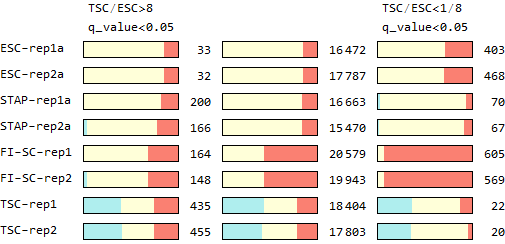
Extraction of SNPs that have TSC-specific alleles
tsSNPs.py by Expo70
import sys
filename = sys.argv[1]
base_pos = { "A":0, "C":1, "G":2, "T":3 }
homozygous_min_ratio = 0.95 #from M&M "SNP identification and heterozygosity tests" in Endo-2014
with open(filename, "r") as fin:
for line in fin:
li = line.rstrip()
ls = li.split("\t")
strains = ls[6-1]
strain_bases = set(list(strains))
acgt = ls[7-1].split(",")
if len(acgt) == 4:
if len(strain_bases) == 1:
common_base = strain_bases.pop()
common_base_count = int(acgt[base_pos[common_base]])
bases = {"A","C","G","T"}
bases.remove(common_base)
for b in bases:
b_count = int(acgt[base_pos[b]])
total_count = common_base_count + b_count
if total_count == 0:
continue
if float(b_count)/total_count >= homozygous_min_ratio:
print li
break
$ cd TSC-rep1/*.tophat
$ python ~/tsSNPs.py snpexp.out | awk '$3!="." {print $3}' | sort -u > SRR1171590.tsSNPs.list
$ cd ../../TSC-rep2/*.tophat
$ python ~/tsSNPs.py snpexp.out | awk '$3!="." {print $3}' | sort -u > SRR1171591.tsSNPs.list
$ cd ../..
$ cat TSC-rep1/*.tophat/SRR1171590.tsSNPs.list TSC-rep2/*.tophat/SRR1171591.tsSNPs.list | sort -u > tsSNPs.list
tsSNPs.list
or common SNPs among TSC replicates
$ comm -1 -2 TSC-rep1/*.tophat/SRR1171590.tsSNPs.list TSC-rep2/*.tophat/SRR1171591.tsSNPs.list > tsSNPs2.listtsSNPs2.list
or common SNPs among TSC replicates including those having no rs IDs
$ python ~/tsSNPs.py snpexp.out | awk '{print $1 "\t" $2}' | sort -u > SRR1171590.tsSNPs3.list
$ comm -1 -2 TSC-rep1/*.tophat/SRR1171590.tsSNPs3.list TSC-rep2/*.tophat/SRR1171591.tsSNPs3.list > tsSNPs3.listtsSNPs3.list in format of "#chr pos"
Whole SNPs analysis of the ratio of heterozygous/homogygous SNPs having TSC-specific alleles
$ grep -Fw -f tsSNPs.list snpexp.out > SRR1171590.tsSNPs.out $ grep -Fw -f tsSNPs2.list snpexp.out > SRR1171590.tsSNPs2.out $ grep -Fw -f tsSNPs3.list snpexp.out > SRR1171590.tsSNPs3.out
tsSNPs-count.py by Expo70
import sys
filename = sys.argv[1]
base_pos = { "A":0, "C":1, "G":2, "T":3 }
homozygous_min_ratio = 0.95
with open(filename, "r") as fin:
nonTsSNPs_homo_count = 0
hetero_count = 0
tsSNPs_homo_count = 0
for line in fin:
li = line.rstrip()
ls = li.split("\t")
strains = ls[6-1]
strain_bases = set(list(strains))
acgt = ls[7-1].split(",")
if len(acgt) == 4:
if len(strain_bases) == 1:
common_base = strain_bases.pop()
common_base_count = int(acgt[base_pos[common_base]])
bases = {"A","C","G","T"}
bases.remove(common_base)
b_counts = list()
for b in bases:
b_counts.append(int(acgt[base_pos[b]]))
m_count = max(b_counts)
if float(common_base_count)/(common_base_count + m_count) >= homozygous_min_ratio:
nonTsSNPs_homo_count = nonTsSNPs_homo_count + 1
elif float(m_count)/(common_base_count + m_count) >= homozygous_min_ratio:
tsSNPs_homo_count = tsSNPs_homo_count + 1
else:
hetero_count = hetero_count + 1
print "# file\tNon-TSC homo\tTSC/non-TSC homo\tTSC-type homo\tTotal #SNPs"
print "%s\t%d\t%d\t%d\t%d" %(filename, nonTsSNPs_homo_count, hetero_count, tsSNPs_homo_count, nonTsSNPs_homo_count + hetero_count + tsSNPs_homo_count)
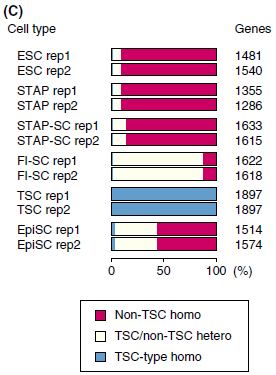
$ for f in $(find -name "*.tsSNPs.out"); do python ~/tsSNPs-count.py $f; done | LC_COLLATE=C sort -uI got
# file Non-TSC homo TSC/non-TSC homo TSC-type homo Total #SNPs ./ESC-rep1a/SRR1171560.tophat/SRR1171560.tsSNPs.out 1324 110 20 1454 ./ESC-rep2a/SRR1171561.tophat/SRR1171561.tsSNPs.out 1385 120 20 1525 ./FI-SC-rep1/SRR1171565.tophat/SRR1171565.tsSNPs.out 285 1339 13 1637 ./FI-SC-rep2/SRR1171566.tophat/SRR1171566.tsSNPs.out 261 1320 10 1591 ./STAP-rep1a/SRR1171580.tophat/SRR1171580.tsSNPs.out 1270 125 14 1409 ./STAP-rep2a/SRR1171581.tophat/SRR1171581.tsSNPs.out 1202 120 16 1338 ./TSC-rep1/SRR1171590.tophat/SRR1171590.tsSNPs.out 0 35 1858 1893 ./TSC-rep2/SRR1171591.tophat/SRR1171591.tsSNPs.out 0 237 1599 1836
$ for f in $(find -name "*.tsSNPs2.out"); do python ~/tsSNPs-count.py $f; done | LC_COLLATE=C sort -uI got
# file Non-TSC homo TSC/non-TSC homo TSC-type homo Total #SNPs ./ESC-rep1a/SRR1171560.tophat/SRR1171560.tsSNPs2.out 1047 82 19 1148 ./ESC-rep2a/SRR1171561.tophat/SRR1171561.tsSNPs2.out 1100 83 18 1201 ./FI-SC-rep1/SRR1171565.tophat/SRR1171565.tsSNPs2.out 187 1076 10 1273 ./FI-SC-rep2/SRR1171566.tophat/SRR1171566.tsSNPs2.out 166 1077 9 1252 ./STAP-rep1a/SRR1171580.tophat/SRR1171580.tsSNPs2.out 1000 87 13 1100 ./STAP-rep2a/SRR1171581.tophat/SRR1171581.tsSNPs2.out 955 82 15 1052 ./TSC-rep1/SRR1171590.tophat/SRR1171590.tsSNPs2.out 0 0 1450 1450 ./TSC-rep2/SRR1171591.tophat/SRR1171591.tsSNPs2.out 0 0 1450 1450
$ for f in $(find -name "*.tsSNPs3.out"); do python ~/tsSNPs-count.py $f; done | LC_COLLATE=C sort -uI got
# file Non-TSC homo TSC/non-TSC homo TSC-type homo Total #SNPs ./ESC-rep1a/SRR1171560.tophat/SRR1171560.tsSNPs3.out 1293 106 38 1437 ./ESC-rep2a/SRR1171561.tophat/SRR1171561.tsSNPs3.out 1357 107 36 1500 ./FI-SC-rep1/SRR1171565.tophat/SRR1171565.tsSNPs3.out 249 1317 20 1586 ./FI-SC-rep2/SRR1171566.tophat/SRR1171566.tsSNPs3.out 224 1318 19 1561 ./STAP-rep1a/SRR1171580.tophat/SRR1171580.tsSNPs3.out 1211 102 29 1342 ./STAP-rep2a/SRR1171581.tophat/SRR1171581.tsSNPs3.out 1157 94 29 1280 ./TSC-rep1/SRR1171590.tophat/SRR1171590.tsSNPs3.out 0 0 1803 1803 ./TSC-rep2/SRR1171591.tophat/SRR1171591.tsSNPs3.out 0 0 1803 1803
Ref: Endo-2014, Fig.4C


Raw outputs
for Fig2B (ESC specific genes)
SRR1171565.fig2B.out (FI-SC-rep1)#Chr Position ID_REF Ref Alt C57BL6NJ,129P2,129S1,129S5 #A,C,G,T Genes
SRR1171566.fig2B.out (FI-SC-rep2)2 168749753 rs33341561 C T CCTTTTTT 0,321,0,4 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168749777 rs32916595 T A TTAAAAAA 0,0,0,334 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168750038 rs33175525 A G AAGGGGGG 172,0,2,0 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168750384 rs33232114 G A GGAAAAAA 3,0,228,0 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168754522 rs32978167 A G AAGGGGGG 274,0,3,0 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168755443 rs13476906 T C TTCCCCCC 0,3,0,190 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 4 55527553 rs49458812 G A GGAAAAAA 6,0,296,0 NM_010637.3 // Klf4 4 55527858 rs52406488 A C AACCCCCC 317,1,0,1 NM_010637.3 // Klf4 4 55527950 rs245328394 A G AAGGGGGG 174,3,2,23 NM_010637.3 // Klf4 4 55528182 rs32168644 A G AAGGGGGG 298,1,5,3 NM_010637.3 // Klf4
SRR1171590.fig2B.out (TSC-rep1)2 168749753 rs33341561 C T CCTTTTTT 1,261,0,3 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168749777 rs32916595 T A TTAAAAAA 5,0,0,292 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168750038 rs33175525 A G AAGGGGGG 175,0,2,0 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168750384 rs33232114 G A GGAAAAAA 2,1,176,0 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168754522 rs32978167 A G AAGGGGGG 257,0,3,0 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168755443 rs13476906 T C TTCCCCCC 0,2,0,202 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 4 55527553 rs49458812 G A GGAAAAAA 10,0,304,0 NM_010637.3 // Klf4 4 55527858 rs52406488 A C AACCCCCC 326,2,0,0 NM_010637.3 // Klf4 4 55527950 rs245328394 A G AAGGGGGG 166,1,0,12 NM_010637.3 // Klf4 4 55528182 rs32168644 A G AAGGGGGG 290,0,4,0 NM_010637.3 // Klf4
SRR1171591.fig2B.out (TSC-rep2)2 168749753 rs33341561 C T CCTTTTTT 0,45,0,24 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168749777 rs32916595 T A TTAAAAAA 24,0,0,48 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168750038 rs33175525 A G AAGGGGGG 32,0,11,0 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168750384 rs33232114 G A GGAAAAAA 20,0,21,0 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168754522 rs32978167 A G AAGGGGGG 28,0,15,0 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168755443 rs13476906 T C TTCCCCCC 0,29,0,23 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 4 55528182 rs32168644 A G AAGGGGGG 0,0,32,0 NM_010637.3 // Klf4
SRR1171560.fig2B.out (ESC-rep1a)2 168749753 rs33341561 C T CCTTTTTT 0,37,0,28 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168749777 rs32916595 T A TTAAAAAA 26,0,0,34 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168750038 rs33175525 A G AAGGGGGG 27,0,13,0 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168750384 rs33232114 G A GGAAAAAA 20,0,27,0 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168754522 rs32978167 A G AAGGGGGG 42,0,22,0 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168755443 rs13476906 T C TTCCCCCC 0,19,0,34 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 4 55527553 rs49458812 G A GGAAAAAA 34,0,0,0 NM_010637.3 // Klf4 4 55528182 rs32168644 A G AAGGGGGG 0,0,58,0 NM_010637.3 // Klf4
SRR1171561.fig2B.out (ESC-rep2a)2 168749753 rs33341561 C T CCTTTTTT 0,107,0,70 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168749777 rs32916595 T A TTAAAAAA 60,0,0,113 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168750038 rs33175525 A G AAGGGGGG 66,0,28,0 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168750384 rs33232114 G A GGAAAAAA 41,0,75,0 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168754522 rs32978167 A G AAGGGGGG 76,0,48,0 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168755443 rs13476906 T C TTCCCCCC 0,48,0,42 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 4 55527553 rs49458812 G A GGAAAAAA 68,1,81,0 NM_010637.3 // Klf4 4 55527858 rs52406488 A C AACCCCCC 81,31,0,0 NM_010637.3 // Klf4 4 55527950 rs245328394 A G AAGGGGGG 54,1,16,5 NM_010637.3 // Klf4 4 55528182 rs32168644 A G AAGGGGGG 58,0,68,2 NM_010637.3 // Klf4
2 168749753 rs33341561 C T CCTTTTTT 0,120,0,80 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168749777 rs32916595 T A TTAAAAAA 74,0,0,109 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168750038 rs33175525 A G AAGGGGGG 39,0,37,0 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168750384 rs33232114 G A GGAAAAAA 63,0,85,0 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168754522 rs32978167 A G AAGGGGGG 90,0,38,0 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 2 168755443 rs13476906 T C TTCCCCCC 0,67,0,73 NM_201396.2 // Sall4 /// NM_201395.2 // Sall4 /// NM_175303.3 // Sall4 4 55527553 rs49458812 G A GGAAAAAA 68,0,93,0 NM_010637.3 // Klf4 4 55527858 rs52406488 A C AACCCCCC 112,45,0,0 NM_010637.3 // Klf4 4 55527950 rs245328394 A G AAGGGGGG 53,2,24,6 NM_010637.3 // Klf4 4 55528182 rs32168644 A G AAGGGGGG 90,0,80,0 NM_010637.3 // Klf4
for Fig2C (TSC specific genes)
SRR1171565.fig2C.out (FI-SC-rep1)#Chr Position ID_REF Ref Alt C57BL6NJ,129P2,129S1,129S5 #A,C,G,T Genes
SRR1171566.fig2C.out (FI-SC-rep2)14 118235390 rs215374979 C T CCTTTTTT 0,29,0,6 NM_177753.3 // Sox21 14 118236534 rs50457356 C T CCTTTTTT 0,37,0,5 NM_177753.3 // Sox21 14 118236794 rs257018347 A G AAGGGGGG 21,0,4,0 NM_177753.3 // Sox21 14 118236818 rs249072519 C T CCTTTTTT 0,20,0,3 NM_177753.3 // Sox21 2 103439276 rs13469469 C T CCTTTTTT 0,52,0,46 NM_001145813.1 // Elf5 /// NM_010125.3 // Elf5 2 103442829 rs13469464 G A GGAAAAAA 40,0,39,0 NM_001145813.1 // Elf5 /// NM_010125.3 // Elf5 2 103449748 rs52190599 C T CCTTTTTT 0,4,0,37 NM_001145813.1 // Elf5 /// NM_010125.3 // Elf5 2 103449764 rs234337412 C T CCTTTTTT 0,2,0,32 NM_001145813.1 // Elf5 /// NM_010125.3 // Elf5 2 103450038 rs13469472 C T CCTTTTTT 0,26,0,12 NM_001145813.1 // Elf5 /// NM_010125.3 // Elf5
SRR1171590.fig2C.out (TSC-rep1)14 118234949 rs253162768 G C GGCCCCCC 0,1,27,0 NM_177753.3 // Sox21 14 118235390 rs215374979 C T CCTTTTTT 0,23,0,7 NM_177753.3 // Sox21 14 118236534 rs50457356 C T CCTTTTTT 0,27,0,0 NM_177753.3 // Sox21 14 118236794 rs257018347 A G AAGGGGGG 23,0,3,0 NM_177753.3 // Sox21 14 118236818 rs249072519 C T CCTTTTTT 0,22,0,3 NM_177753.3 // Sox21 2 103439276 rs13469469 C T CCTTTTTT 0,50,0,46 NM_001145813.1 // Elf5 /// NM_010125.3 // Elf5 2 103442829 rs13469464 G A GGAAAAAA 40,0,45,0 NM_001145813.1 // Elf5 /// NM_010125.3 // Elf5 2 103449748 rs52190599 C T CCTTTTTT 0,2,0,30 NM_001145813.1 // Elf5 /// NM_010125.3 // Elf5 2 103449764 rs234337412 C T CCTTTTTT 0,0,1,28 NM_001145813.1 // Elf5 /// NM_010125.3 // Elf5 2 103450038 rs13469472 C T CCTTTTTT 0,13,0,19 NM_001145813.1 // Elf5 /// NM_010125.3 // Elf5
SRR1171591.fig2C.out (TSC-rep2)14 118234949 rs253162768 G C GGCCCCCC 0,15,9,0 NM_177753.3 // Sox21 14 118235390 rs215374979 C T CCTTTTTT 0,19,0,22 NM_177753.3 // Sox21 14 118236534 rs50457356 C T CCTTTTTT 0,9,0,28 NM_177753.3 // Sox21 14 118236794 rs257018347 A G AAGGGGGG 18,0,15,0 NM_177753.3 // Sox21 14 118236818 rs249072519 C T CCTTTTTT 0,15,0,13 NM_177753.3 // Sox21 2 103439276 rs13469469 C T CCTTTTTT 1,429,3,460 NM_001145813.1 // Elf5 /// NM_010125.3 // Elf5 2 103442829 rs13469464 G A GGAAAAAA 305,0,362,1 NM_001145813.1 // Elf5 /// NM_010125.3 // Elf5 2 103449748 rs52190599 C T CCTTTTTT 0,36,1,277 NM_001145813.1 // Elf5 /// NM_010125.3 // Elf5 2 103449764 rs234337412 C T CCTTTTTT 0,19,0,255 NM_001145813.1 // Elf5 /// NM_010125.3 // Elf5 2 103450038 rs13469472 C T CCTTTTTT 0,152,0,138 NM_001145813.1 // Elf5 /// NM_010125.3 // Elf5
SRR1171560.fig2C.out (ESC-rep1a)14 118234949 rs253162768 G C GGCCCCCC 0,10,13,0 NM_177753.3 // Sox21 14 118235390 rs215374979 C T CCTTTTTT 0,26,0,23 NM_177753.3 // Sox21 14 118236534 rs50457356 C T CCTTTTTT 0,16,0,37 NM_177753.3 // Sox21 14 118236794 rs257018347 A G AAGGGGGG 12,0,17,0 NM_177753.3 // Sox21 14 118236818 rs249072519 C T CCTTTTTT 0,12,0,18 NM_177753.3 // Sox21 2 103439276 rs13469469 C T CCTTTTTT 1,371,0,478 NM_001145813.1 // Elf5 /// NM_010125.3 // Elf5 2 103442829 rs13469464 G A GGAAAAAA 299,0,322,0 NM_001145813.1 // Elf5 /// NM_010125.3 // Elf5 2 103449748 rs52190599 C T CCTTTTTT 0,44,1,255 NM_001145813.1 // Elf5 /// NM_010125.3 // Elf5 2 103449764 rs234337412 C T CCTTTTTT 0,22,0,228 NM_001145813.1 // Elf5 /// NM_010125.3 // Elf5 2 103450038 rs13469472 C T CCTTTTTT 0,145,0,114 NM_001145813.1 // Elf5 /// NM_010125.3 // Elf5
SRR1171561.fig2C.out (ESC-rep2a)
Reference
- Endo 2014, Genes to Cells, "Quality control method for RNA-seq using single nucleotide polymorphism allele frequency"
- Katsura report (J)(E)