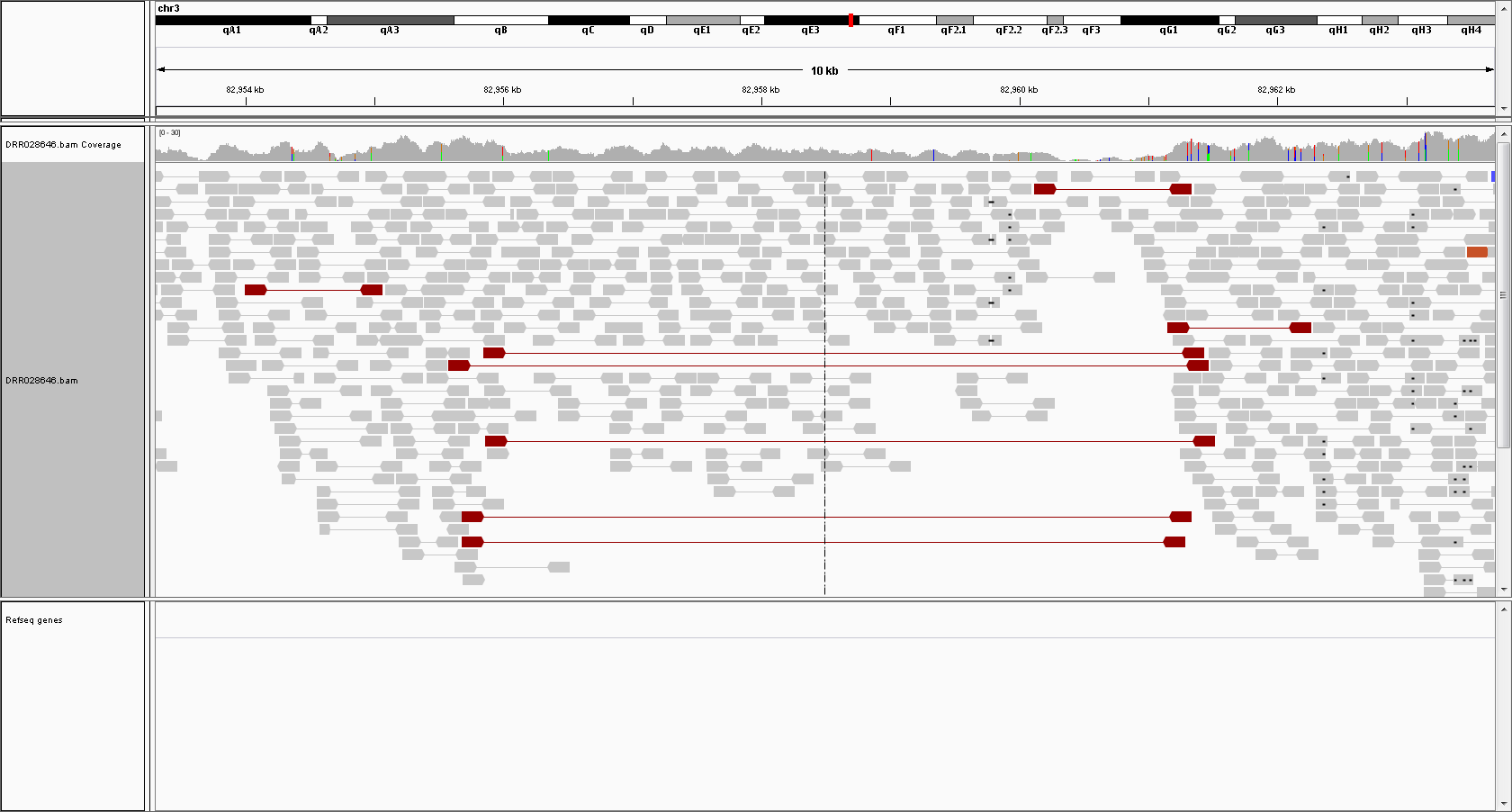
Deletions
5kb deletion (hetero) in Chromosome 3
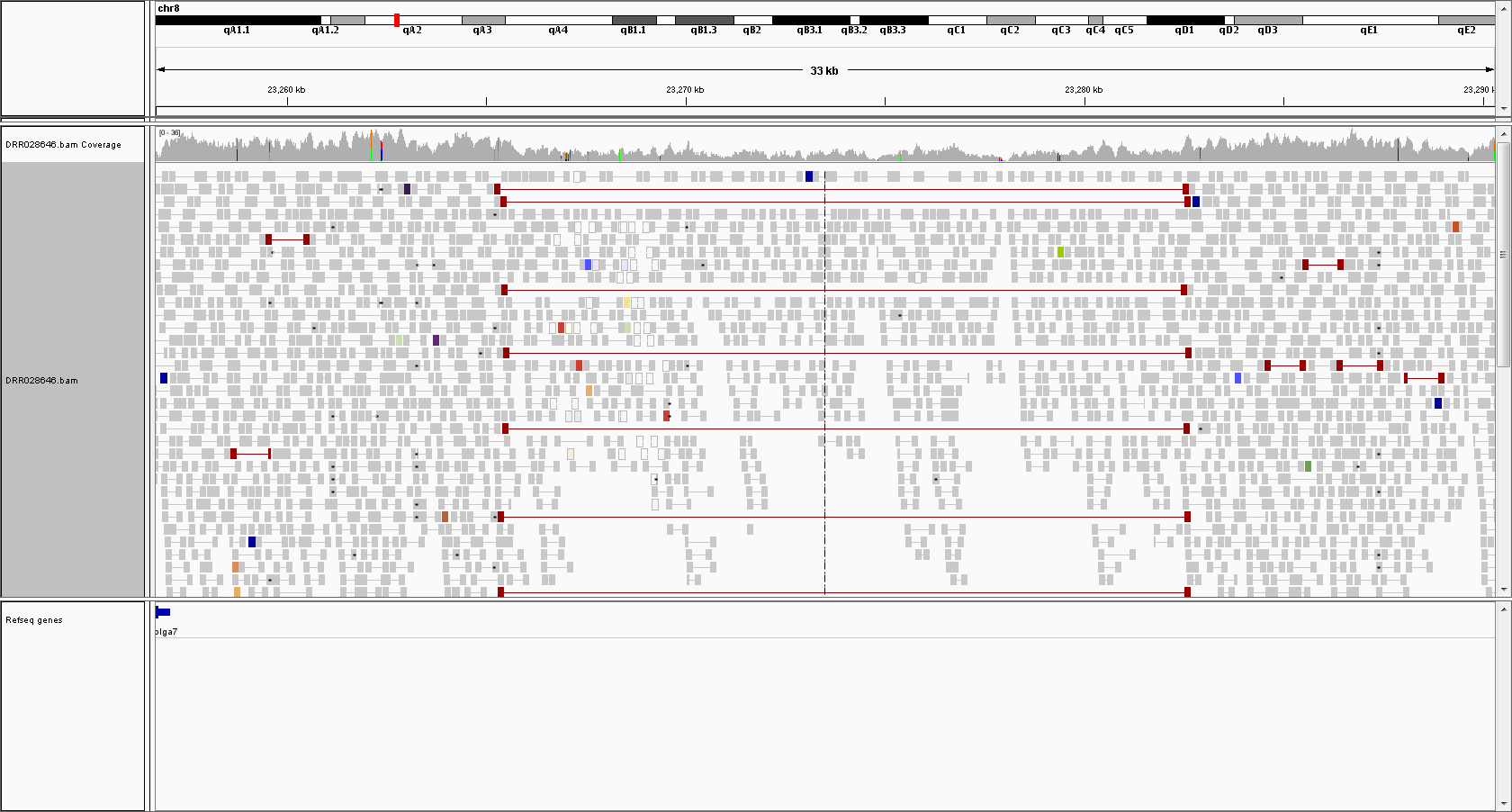
17kb deletion (hetero) in Chromosome 8
Transgene related
Acrosin
Large coverage in upstream and a part of its coding region indicates that this region is used as a transgene and multiple copies are inserted in the genome.

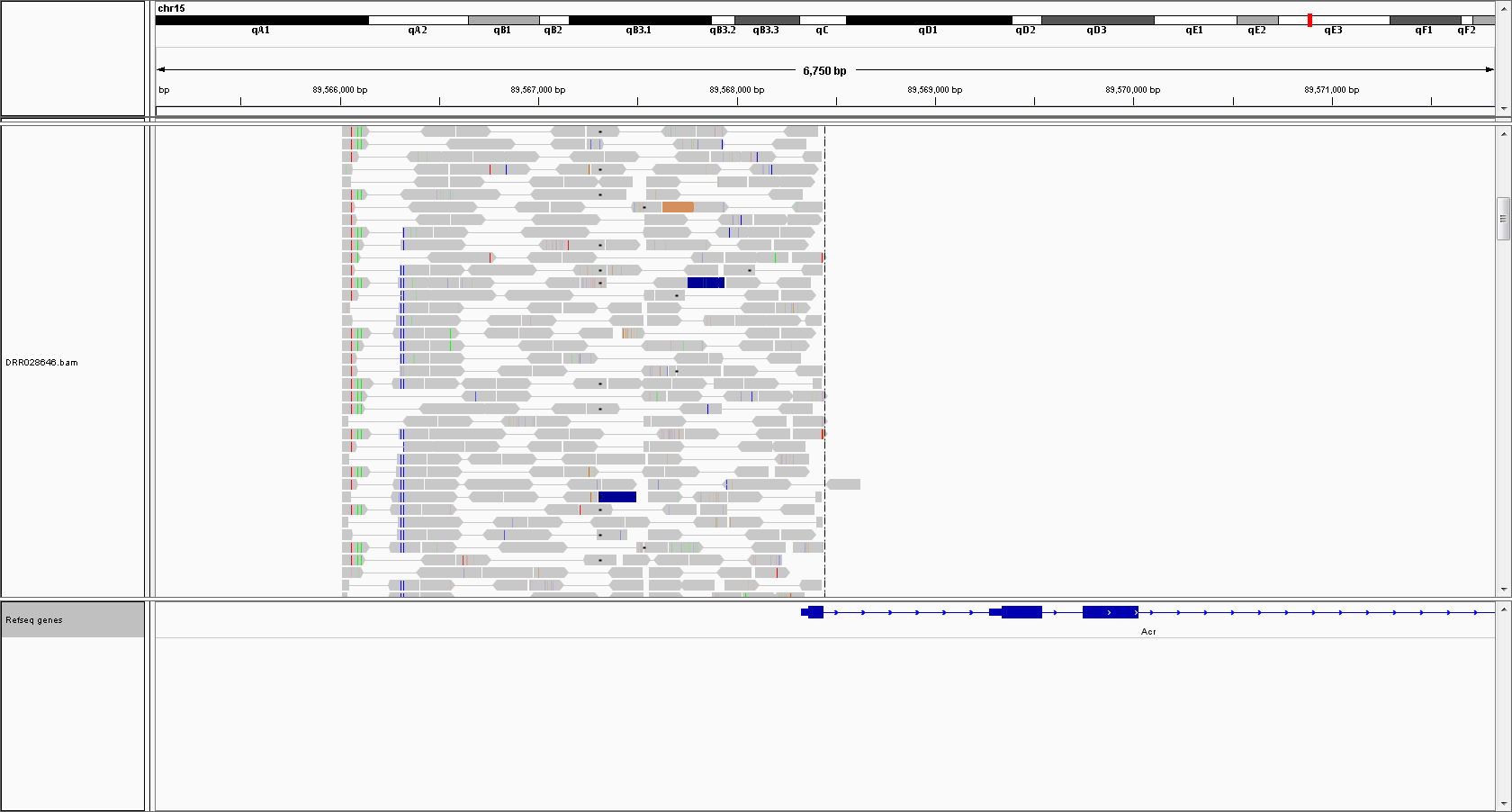
Mean coverage in this 2.4-kb region was 108.4.
$ samtools mpileup -f ../../genome/Mus_musculus.GRCm38.74.dna.toplevel.fa.gz DRR028646.bam -r15:89566016-89568433 \
$ awk '{sum=+$4} END {print sum/NR}'
Mean coverages of the flanking 2.4-kb regions of Acrsin promoter region was 11.4 and 11.1. Genome has 2 copies of acrosin promoter region and thus exptected coverage of the 2.4-kb region relative to its surrounding region is (n + 2)/2, where n is the copy number of acrosin promoters in the inserted transgenes. From this model, I can estimate n = 17.
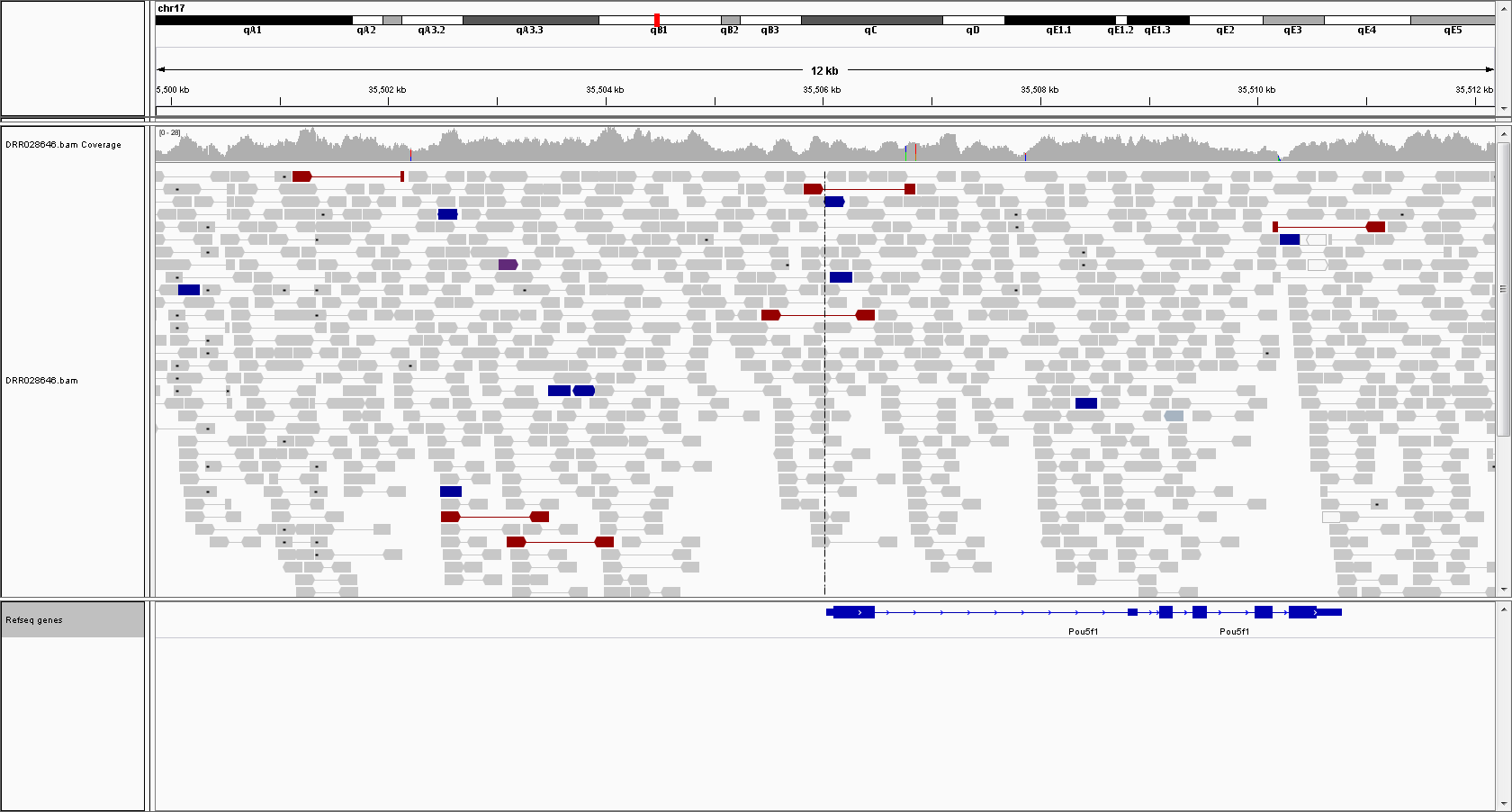
Oct4
There's no indication of the use of Oct4 promoter as a transgene in thie genome.