
I have found a sequence structure below in FES1 genome so far.

$ bwa index -p egfp_index egfp.fasta $ bwa mem egfp_index DRR028646_1.fasta | grep EGFP > DRR028646_1_egfp.sam $ bwa index -p caggs_index caggs.fasta $ bwa mem caggs_index DRR028646_1.fasta | grep caggs > DRR028646_1_caggs.sam $ bwa index -p rabbit-pA_index rabbit-pA.fasta $ bwa mem rabbit-pA_index DRR028646_1.fasta | grep rabbit-pA > DRR028646_1_rabbit-pA.sam $ $ samtools view -bS rabbit-pA.sam > rabbit-pA.bam $ #v0.1.19 $ samtools sort rabbit-pA.bam rabbit-pA $ #v1.3.1, and use .sort.bam file instead of .bam file $ samtools sort rabbit-pA.bam -o rabbit-pA.sort.bam $ samtools index rabbit-pA.bam $ samtools tview rabbit-pA.bam ../../transgene/rabbit-pA.fasta $ $ samtools mpileup -uf ../../transgene/rabbit-pA.fasta rabbit-pA.bam | bcftools view -vcg - > rabbit-pA.vcfalignment on this tail-to-tail structure
| transgene | coverage |
|---|---|
| EGFP | 76.6 |
| CAG | 21.6 |
| acrosin promoter | 138.0 |
| rabbit globin pA | 121.3 |
in mouse chromosome 3, position 37732943

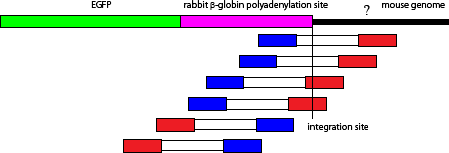
I found this site by mapping mate pair reads of the reads that were aligned on rabbit globin pA sequence as below.
$ samtools view -h DRR028646_1_rabbit-pA.bam | grep -v "^@" | cut -f 1 > DRR028646_1_rabbit-pA.read_id $ sed -e "s/^/^@/" -e "s/$/\\\s/" DRR028646_1_rabbit-pA.read_id > DRR028646_1_rabbit-pA.read_id.pattern $ zgrep -A3 -f DRR028646_1_rabbit-pA.read_id.pattern DRR028646_2.fastq.gz > DRR028646_2_1-rabbit-pA.fastq $ grep -v "^--" DRR028646_2_1-rabbit-pA.fastq > temp $ mv temp DRR028646_2_1-rabbit-pA.fastq $ bwa mem ../../genome/GRCm38_index DRR028646_2_1-rabbit-pA.fastq > DRR028646_2_1-rabbit-pA.sam $ samtools view -bS DRR028646_2_1-rabbit-pA.sam > DRR028646_2_1-rabbit-pA.bam $ samtools sort DRR028646_2_1-rabbit-pA.bam DRR028646_2_1-rabbit-pA $ samtools index DRR028646_2_1-rabbit-pA.bam $ samtools tview DRR028646_2_1-rabbit-pA.bam ../../genome/Mus_musculus.GRCm38.74.dna.toplevel.fa
then do the command g and type 3:37733000.
I found that two reads from DRR028646_1.fastq contain the boundery between rabbit polyA sequence and mouse chromosome 3.
DRR028646.25410863 (reverse complement)CTAGCCAGATTTTTCCTCCTCTCCTGACTACTCCCAGTCATAGCTGTCCCTCTTC|CACTGTGCCCGTGGGTGCTGGGGTGGAAGGGCCATGCTTGTCCAGAGCACAATTCCTCCACCAAAGCTATGCCTTTCCGAATGACTTCTGCCTTATDRR028646.35838511 (reverse complement)
TTTTTATGTTTTGTTTTGTGTTATTTTTTTCTTTAACATCCCTAAAATTTTCCTTACATGTTTTACTAGCCAGATTTTTCCTCATCTCCTGACTACTCCCAGTCATAGCTGTCCCTCTTC|CACTGTGCCCGTGGGTGCTGGGGTGGAAGGG
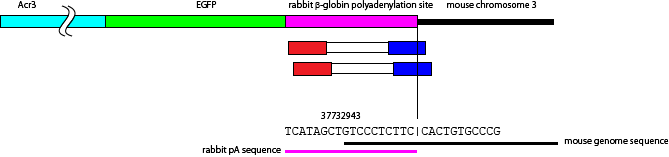
Some of the other reads have HindIII-BamHI region from rabbit pA sequence from pCAGGS. But above two sequence have no such region. Thus I speculated that the right end (on the reference genome) of transgene(s) in the chromosome 3 is Acr3-GFP.
I searched the other end of the integration site. From the analysis (1) and (2), I know an integration site reside around 3:37732943. Next I searched reads that are aligned on left 1kb region of the integration site, i.e. 3:37731943-37732943.
$ samtools faidx ../../genome/Mus_musculus.GRCm38.74.dna.toplevel.fa 3:37731943-37732943 > ../../genome/chr3_37731943-37732943.fasta
$ samtools index -p ../../genome/chr3_37731943-37732943_index ../../genome/chr3_37731943-37732943.fasta
... not found