SNP analysis of the genome of STAP related cells
In the genome of 129B6 F1 mouse, one chromosome is derived from 129 parent, and the other is derived from B6 parent. Actually, you can find most of the SNVs are found in around 50% of the reads covering those sites when you see the alignment results in a viewer like IGV, indicating that these SNVs are hetrozygous. As the reference genome is from B6 mouse, SNVs found in .vcf file is thought to be the difference between 129 and B6 alleles but they also include a few point mutations that occur in B6 chromosome.
How can you determine whether a SNV is 129 allele or it is a mutation in B6 chromosome? Authors of the original analysis of STAP related cells tackled this problem by using a SNPs database of laboratory mouse strains (Mouse Genomes Project). The authors extracted 129-stain specific SNPs as a set of SNPs that were commonly found in three 129-related strains but not fund in a B6-related stain in the database. I followed the authors' method as below although there were no specific ways of doing this extraction described in the paper.
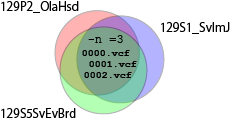
$ mkdir isec1 $ bcftools isec 129P2_OlaHsd.mgp.v5.snps.dbSNP142.vcf.gz 129S1_SvImJ.mgp.v5.snps.dbSNP142.vcf.gz 129S5SvEvBrd.mgp.v5.snps.dbSNP142.vcf.gz -n =3 -p isec1 -OzI obtained 5073728 SNPs that are common to all the 129 strains.
$ mkdir isec2 $ bcftools isec isec1/0000.vcf.gz C57BL_6NJ.mgp.v5.snps.dbSNP142.vcf.gz -C -p isec2 -OzI finally obtained 5041214 SNPs that are common to all the 129 strains but not observed in B6NJ strain.
$ cp isec2/0000.vcf.gz 129SNPs.vcf.gz $ bcftools index 129SNPs.vcf.gz
analysis (1)
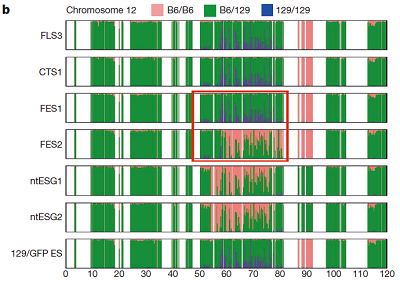
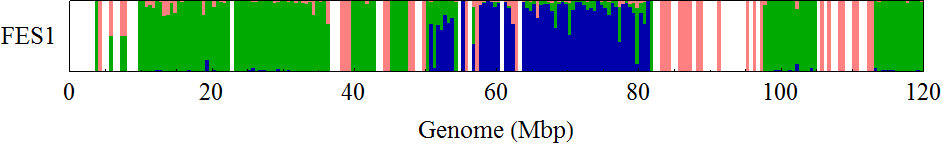
FES1 Chromosome 12
Ref: Konno et al., 2015, Fig 1b.
$ mkdir isec $ bcftools isec DRR028646.vcf.gz ../../snps/129SNPs.vcf.gz -p isec -Oz $ cd isecIn
README.txt created by this command, you can see which file is which group intersection:
This file was produced by vcfisec. The command line was: bcftools isec -p isec -Oz DRR028646.vcf.gz ../../snps/129SNPs.vcf.gz Using the following file names: isec/0000.vcf.gz for records private to DRR028646.vcf.gz isec/0001.vcf.gz for records private to ../../snps/129SNPs.vcf.gz isec/0002.vcf.gz for records from DRR028646.vcf.gz shared by both DRR028646.vcf.gz ../../snps/129SNPs.vcf.gz isec/0003.vcf.gz for records from ../../snps/129SNPs.vcf.gz shared by both DRR028646.vcf.gz ../../snps/129SNPs.vcf.gz
$ zcat 0000.vcf.gz | awk '{if(length($4)==1 && length($5)==1) print $1 "\t" $2 "\t" $6 "\t" $10}' > DRR028646-B6SNP.tsv
$ zcat 0002.vcf.gz | awk '{if(length($4)==1 && length($5)==1) print $1 "\t" $2 "\t" $6 "\t" $10}' > DRR028646-129SNP.tsv
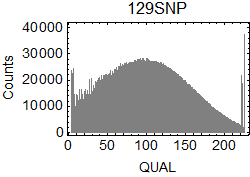
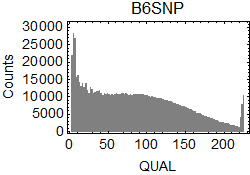
I extracted SNPs from the results of bcftools isec. In .vcf files, I focused on the columns of CHROM ($1), POS ($2), REF ($4), ALT ($5), QUAL ($6), and INFO/GT ($10) in the .vcf files.
![]()
I drew the ratio plot above on Windows by using a script language. I only counted the SNPs with high reliability (QUAL > 200.0). The bin size of the counting was 0.5Mbp. Because I didn't know how authors of the original paper found B6/B6 sites, I counted sites in B6SNP.tsv that have GT = 1/1 as B6/B6, sites in 129SNP.tsv that have GT = 0/1 as B6/129, and sites in 129SNP.tsv that have GT = 1/1 as 129/129 in this analysis (1).
The obtained distribution of SNPs was somehow different from that in the original paper. But it captured the unique mosaic sturucture of 129/129-type SNPs distributions in homologous chromosomes 6, 11, and 12, which was similar to that observed in the original paper.
QUAL scores of the extracted SNPs: