129X1/SvJ - SRR1614029
129X1/SvJ is a substrain of an inbred mice 129. It was originally called as 129/SvJ, but after it was found to be genetically contamined by an unknown stain, it was renamed as 129X1/SvJ. To obtain its genomic sequence, I serached DDBJ database for 129X1/SvJ whole genome sequencing (WGS) by using DRASearch (Keyword: "129X1/SvJ"). I found a submission SRA191208, which contains SRS722749 (Low coverage sequencing of 129X1/SvJ).
obtaining SRA toolkit
Its sequencing data is provided only in SRA format (a compressed archive).$ wget -c https://ftp-trace.ncbi.nlm.nih.gov/sra/sdk/2.8.1-2/sratoolkit.2.8.1-2-ubuntu64.tar.gz $ tar -xzvf sratoolkit.2.8.1-2-ubuntu64.tar.gz
data retrieval and QC
$ wget -c ftp://ftp.ddbj.nig.ac.jp/ddbj_database/dra/sralite/ByExp/litesra/SRX/SRX734/SRX734252/SRR1614029/SRR1614029.sra $ ~/sratoolkit.2.8.1-2-ubuntu64/bin/fastq-dump SRR1614029.sra $ rm SRR1614029.sra $ mdkir reports $ nohup ~/FastQC/fastqc -o reports -t 1 SRR1614029.fastq &The original SRA file (0.4G byte) was converted to a fastq file (3.5G byte). I obtained the FastQC result.
alignment
$ nohup bwa aln -n0.02 ../../genome/GRCm38_index SRR1614029.fastq > SRR1614029.sai 2>>log.txt & $ nohup bwa samse ../../genome/GRCm38_index SRR1614029.sai SRR1614029.fastq 2> stderr.log | samtools view -bS - > SRR1614029.bam 2> stderr.2.log & $ nohup samtools sort SRR1614029.bam -o SRR1614029.sort.bam & $ mv SRR1614029.sort.bam SRR1614029.bam $ samtools index SRR1614029.bam
genome-wide SNPs distribution analysis
Mouse Phenome Database at The Jackson Laboratory is a useful source of information about mouse strain diversity.
As the result of a query for CGD-MDA1 dataset, you can see mutual differences among some of the inbred 129 starains and the reference C57B6/6J (*).
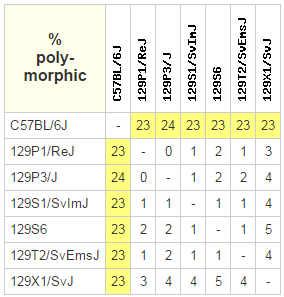 based on 470822 SNPs
based on 470822 SNPs
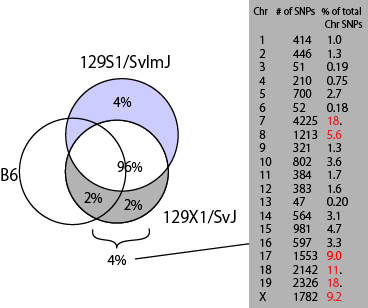
The result indicated that 129X1/SvJ is significantly different from the other 129 substrains in the database.
This has been already known in the literature that...
*This analysis was originally done in a blog comment by "Zaibei PostDoc" and I replicated the result here.
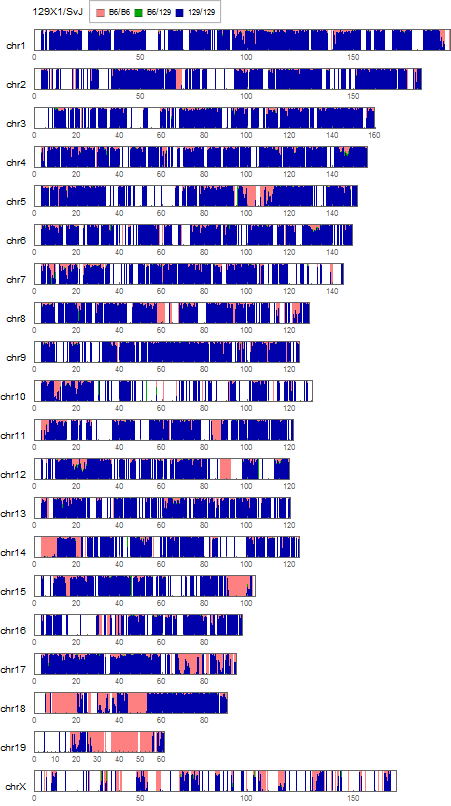
To visualize its genome-wide heterogeneity, I calculated the positional distribution of the allele frequencies of "129"-type and "B6"-type SNPs by using a technique used before. In previous analyses, "129"-type was determined based on the nucleotide bases of three 129 substarains: 129P2/OlaHsd, 129S1/SvImJ, and 129S5SvEvBrd. Here I use them to detect some of the 129X1/SvJ-specific SNPs compared to other three substrains as "B6"-type ones.
$ ~/snpexp/src/snpexp2 -V ~/stap/snps/dbSNP/129B6-SNPs.vcf SRR1614029.bam > SRR1614029.129B6.snpexp2.out
visualization parameters:
- bin size = 0.25Mb
- for each SNP,
- B6/B6 homo: B6 type allele > 95%
- 129/129 homo: B6 type allele < 5%
- 129/B6 hetero: the others